

Significance for the Modification and Origin of Genes
GORDON C. MILLS
Dept. of Human Biological Chemistry &
Genetics
Univ. of Texas, Medical Branch
Galveston, TX 77555
From: Perspectives on Science and Christian Faith 44 (December 1992): 236-245.
The author presents data on the structure of cytochrome c genes in various organisms including the gene for a tissue-specific (testes) cytochrome c. Nucleotide sequences in introns and in noncoding regions of the gene that precede and follow the coding region are discussed, along with regulatory sequences found in these regions. The possible role of design and chance is considered in discussing the origin of the testicular cytochrome c gene and in the origin of pseudogenes. The evidence is reviewed for homology of the various functional cytochrome c genes. Also discussed is the role of presuppositions in regard to hypotheses concerning the origin of informational molecules.
In this paper, I will describe recent studies on genes of cytochrome c, and will attempt to evaluate the significance of these studies on our understanding of evolutionary changes in cytochrome c proteins and in cytochrome c genes. In addition, I will discuss possible homologies of cytochrome c molecules and of the genes that code for and control synthesis of cytochrome c proteins.
Initially, I wish to note a 1987 paper proposing a more stringent usage of the words "homology" and "similarity." The following are quotations from that paper which was authored by eleven researchers in the field of molecular evolution:
Homology should mean "possessing a common evolutionary origin"....Evidence for homology should be explicitly laid out .... Sequence similarities (or other types of similarity) should simply be called "similarities"....Homology among similar structures is a hypothesis that may be correct or mistaken, but a similarity itself is a fact, however it is interpreted.1
Since the meaning of these words is very important in making interpretations, I shall use the words "homology" and "similarity" in the sense agreed on by these eleven authors.
Amino acid sequences in cytochrome c protein molecules have
been determined in about ninety different animal species. Similarities in these
sequences have been used as a primary basis for determining relationships of
these animals to one another. In many instances, (e.g., many high school biology
textbooks) the similarities are cited as a major argument for ancestral
relationships among various classes and phyla of organisms. Since the
information for the sequence of amino acids in a protein molecule resides in the
coding region* of the corresponding gene, in this paper I will review recent
studies on nucleotide sequences in cytochrome c genes.
* See glossary on p. 245 for definitions of
this and other selected terms.
There are several distinctive differences that should be noted when we study sequences in genes instead of in proteins. First, it takes three nucleotides in the gene to code for one amino acid. Hence, for the linear sequence of 104 amino acids in human cytochrome c, the corresponding coding region of the gene would have a linear sequence of 312 nucleotides or 104 codons, with three nucleotides per codon. Secondly, a change in a coding region nucleotide (particularly in the third position of a codon) does not always cause a change in amino acids in the protein molecule. The reason for this is that there are 64 possible three-base arrangements of the four different nucleotides found in DNA coding regions, and only 20 different amino acids are found in proteins. Although some codons are used as start signals (initiation codons) or stop signals (termination codons), this means that there is more than one codon per amino acid. Thirdly, there are portions of the cytochrome c gene at the 5'-end and at the 3'-end of the coding region that are not involved in determining the amino acid sequence of the protein, but instead act by determining whether or not any protein is produced. Certain modifications in these control or regulatory regions may mean that no protein is produced, while other modifications may lead to a decrease in rate, or possibly to an increase in rate of protein formation. It is becoming increasingly evident that these control regions of a gene are just as important as the coding region of the gene.
What is the special significance of studies of nucleotide sequences in cytochrome c genes? First of all, these studies extend sequence comparisons beyond coding regions to portions of genes that control expression of coding regions. Also, studies of other cytochrome c-like genes (pseudogenes) introduces a whole new and unknown element. Are these apparently nonfunctional cytochrome c-like genes remnants of the past evolutionary history of an organism? Or are they fragments of genes that are retained in the genome for possible incorporation into some new or different gene? Or are they just "junk" that will ultimately be broken down and eliminated from the organism? In this paper, the author will present and evaluate data related to the above problems, and suggest how certain presuppositions might have a role in our explanations and in formulating hypotheses.2
Cytochrome c Structure and Function
Cytochrome c functions in a respiratory chain in cells by virtue of having a heme prosthetic group. The central iron atom of the heme undergoes reversible oxidation and reduction during aerobic respiration. In mammals, the cytochrome c protein is made up of a linear chain of 104 amino acids, while in other eukaryotic organisms (organisms whose cells have a nucleus), the chain length ranges from 103 to 112 amino acids. Cytochrome c has a relatively fixed three-dimensional structure with fourteen different amino acids packed tightly around the heme. Several precisely defined channels permit the flow of electrons from the exterior to and from the heme iron. There are 21 amino acid positions in the cytochrome c molecule that are invariant (i.e., if they are replaced, enzymic function is lost). At approximately 20 other positions, the amino acid can be replaced only by one or two very similar amino acids.3 Thus the picture we have of a cytochrome c molecule is one of a highly restricted three-dimensional structure with only limited possibilities for variation. This is consistent with conclusions made by the author in previous articles regarding the structure of hemoglobin4,5 and of aminoacyl tRNA synthetases.6
Cytochrome c Gene Structure
The genetic information of cells (made up of many genes) is stored in the DNA of chromosomes of the cell nucleus. To be expressed, that information is first copied into RNA (the process of transcription), and then is changed into sequences of amino acids in a protein molecule (the process of translation). Each of these processes is very complex, and I will not consider them in detail. It is never easy to determine where in the DNA of cells a gene begins or where it ends. The enzymes (restriction endonucleases) that are utilized to cleave huge DNA molecules into fragments amenable to study do not select sites for cleavage at the beginning or end of genes. Consequently, a number of different types of study, including the study of messenger RNA (mRNA) molecules, are necessary before one decides what makes up a particular gene.
Cytochrome c genes have now been sequenced in rat, mouse, chicken, human, fruit fly and yeast, largely due to studies by Wu and co-workers7-10 and by Scarpulla and co-workers.11-13 The techniques for isolation and sequencing of these genes are difficult and beyond the scope of this presentation, but great credit is due the researchers named above for their careful experimental work. The structure of the rat somatic cytochrome c gene (cyto cs), which is found in all cells, and the gene of a cytochrome c isozyme found only in testes (cyto ct) are shown schematically in Figure 1. It will be noted that there is an intervening nucleotide sequence (intron) in the
|
|
middle of the coding region of the gene. Also, there are long stretches of noncoding nucleotide sequences at both the 5'- and 3'-ends of the coding region of the gene, with additional intron(s) in the 5'-noncoding region. All introns are precisely removed from the nucleotide sequence after transcription during the processing of mRNA. Consequently, mRNA that is utilized for protein synthesis has two separate coding regions joined together with no intron separating the two portions. Within noncoding regions of cytochrome c genes, there are short sequences of nucleotides that serve to control expression of the coding region (that is, to control translation of genetic information from nucleotide sequences to amino acid sequences). A great deal of research is presently underway in order to identify these various control or regulatory sequences.14 Although I have shown a schematic representation of the cyto ct gene as well as the cyto cs gene in Figure 1, I will simply note at this point that there are marked differences in structure of these two genes. The significance of these differences will be considered in more detail later.
In Table 1, I have summarized some differences noted in cytochrome c genes of various species that have been studied. Note that introns are found in mammalian cytochrome c genes and in the genes of chickens, but introns
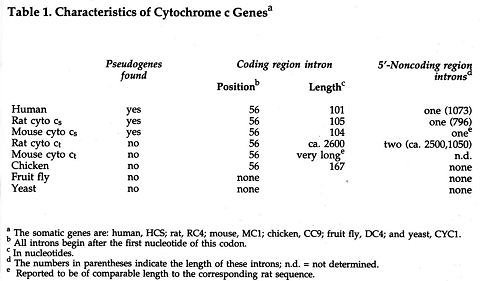 |
are not found in the cytochrome c genes of fruit flies or yeast. It is interesting to note that the position of the coding region intron is the same in all cases (after the first position of codon 56), even though the length of the intron varies. Although the precise function of introns is not known, some control sequences have also been found in introns.
It may also be seen in Table 1 that some cytochrome c-like genes, termed "processed pseudogenes," have been found in the different mammals that were studied, but not in chickens, fruit flies or yeast. The pseudogenes in rats or mice are clearly related to cyto cs genes, but not to cyto ct genes. Processed pseudogenes do not have introns and they have lost various control sequences in noncoding regions. In coding regions, the degree of difference of these pseudogenes from the functional cytochrome c gene varies markedly. In Figure 2, 13-nucleotide segments
 |
 |
of nine different human pseudogenes are shown. This figure and the data in Table 2 illustrate the type of changes that occur. Some pseudogenes have extensive deletions or insertions of nucleotides and many nucleotide changes. Other pseudogenes have only a few nucleotide changes, and one pseudogene (RC9 of the rat) has the correct rat cytochrome c gene coding sequence. Nevertheless, these pseudogenes are all defective in some manner and are not used for production of functional cytochrome c molecules.
Probably the best explanation of the origin of these pseudogenes is that they were incorporated into DNA initially by reverse transcription of cytochrome c mRNA. This would account for their lack of introns. Once present in DNA, pseudogenes would be copied during cell division and hence would be passed from one generation of cells to the next. In order to be transmitted to subsequent generations of animals, pseudogenes would have to be generated either during formation of ova or sperm or of precursors of these cells. At present, 20 different cytochrome c-like pseudogenes have been studied (11 in humans, 6 in rats, and 3 in mice). None of these pseudogenes are identical.
The Tissue-Specific Isozyme of Cytochrome c (Cyto c1)
The report of a tissue-specific isozyme of cytochrome c in 1975 presented the possibility of some new and interesting aspects of cytochrome c studies. This finding opens again the question of the manner of origin of new enzymes or isozymes as one proceeds from lower to higher forms of life. The traditional answer to this question, at least for isozymes with appreciable sequence and structural similarity, has been to postulate a gene duplication, after which each of the two duplicate genes changes independently with time as a consequence of mutations, gene conversions, etc. I have examined the experimental evidence to see whether the above explanation may be applied to the formation of cyto ct, the testicular isozyme of cytochrome c.15 It should be noted, however, that nucleotide sequence studies at present have been made of cyto ct only in rats and in mice.16 Immunologic studies have shown the presence of cyto ct in two other species (rabbit and bull), but the presence of cyto ct has not been demonstrated in humans.17
An examination of amino acid sequence data for cyto ct and a comparison of it with cyto cs data presents some very striking differences. Rat cyto ct differs from rat cyto cs in 15 positions of the amino acid sequence of the protein, while mouse cyto ct differs from mouse cyto cs in 14 positions. Rat and mouse cyto ct differ in 4 positions, whereas rat and mouse cyto cs proteins are identical. The differences between cyto ct and cyto cs proteins do not involve invariant amino acids; in fact, most of these differences involve amino acids on the exterior of the three-dimensional molecule. However, many of the amino acid differences are of a radical nature. By radical, I mean that the nature of the R-group of the amino acid has been markedly changed (that is, a hydrophobic group for a hydrophylic group; a charged group for a neutral group, etc.) Also, amino acid differences in cyto ct of either rat or mouse are not commonly seen when one examines other cytochrome c sequences. Thus, the experimental evidence involving amino acid sequence data indicates that cyto ct is quite divergent from somatic cytochrome c of all other organisms, and can not be readily related to somatic cytochromes c in any postulated phylogenetic tree.18
A nucleotide difference matrix for the coding region of cytochrome c genes is shown in Figure 3. In this matrix I have provided data not only for the gene of the primary cytochrome c of cells, but also for the gene of the
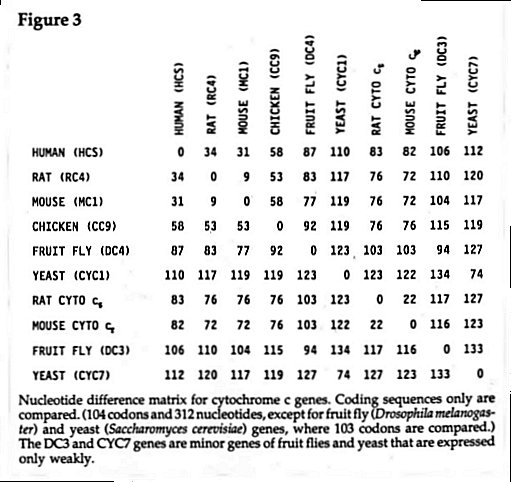 |
tissue-specific isozyme of rats and mice (cyto ct) and for the minor cytochrome c genes of fruit flies (DC3) and yeast (CYC7). Thus these studies on coding region sequences are in agreement with the studies involving amino acid sequences; namely, that there is a large divergence of the cyto ct isozymes from the somatic cytochromes c. In addition, from the time of divergence of rats and mice, genes for cyto ct isozymes in these two rodents have diverged much more than genes for the corresponding cyto cs isozymes.19
If we examine noncoding regions of the cyto ct gene of rats, no similarity is found with corresponding positions of the cyto cs gene of rats. This means that if genes of these two cytochrome c isozymes did arise by a gene duplication, then all controlling sequences in these noncoding regions have been replaced during the evolutionary history of the gene. This indicates that genes of the cyto ct isozymes are even more different from the corresponding cyto cs genes than would have been suggested by examining only the coding region nucleotide sequences. If indeed the genes for these two cytochrome c isozymes did arise by gene duplication, is there some built-in control mechanism (possibly subject to chance events) that could be responsible for subsequent changes? Such a built-in control mechanism would, when triggered, lead to the replacement of the noncoding region controls that are appropriate for a somatic enzyme with new controls that are necessary for a gene that functions only in a single tissue (that is, in sperm cells of testes). In other words, is the origin and function of the cyto ct isozyme a consequence of the initial design built into the genes (although possibly triggered by chance events), or is the origin of the new isozyme entirely subject to a sequence of chance events (that is, one chance event after another leading to the replacement of all the necessary nucleotides in the coding region and the replacement of all the necessary control regions in the noncoding regions of the genes)? At present, we do not have answers to these questions, but it is clear that one's initial presuppositions regarding origins may have a role in the development of hypotheses to be utilized in guiding subsequent laboratory investigation in this area.20
Significance of Processed Pseudogenes
As mentioned previously, twenty different cytochrome c-like pseudogenes have been studied in mice, rats and humans. Evans and Scarpulla21 noted that nine of the eleven pseudogenes in humans appeared to be more like the functional rodent somatic cytochrome c genes than they were like the functional human cytochrome c gene (HCS gene). They based their conclusion on similarities that were readily evident (see codons 11, 12, 15 and 20 of Figure 2). These authors indicated that these nine human pseudogenes (designated HC pseudogenes) had much greater similarity to a consensus of nonprimate mammalian cyto cs genes, and consequently would have remained as a nonfunctional part of the mammalian genome for 25 million or more years. In effect, this means that there are molecular remnants remaining in an organism that may reflect some of the past molecular history of the genome of that organism. I have examined the data of Evans and Scarpulla22 carefully to see if their interpretation is clearly supported by the data. To do this, I have compared coding region sequence data for these nine pseudogenes with sequence data for the human HCS gene and the mouse MC1 gene, both of which are functional. Data in Figure 4 are for all positions where nucleotides of the MC1 gene differ from those in the HCS gene. As seen in Figure 4, there are indeed some portions of these nine human pseudogenes (for example, codons 10-22 and 44-51) that
 |
more nearly resemble the mouse gene. However, more than half the length of each of the pseudogenes more closely resembles the human gene. This suggests that portions (especially codons 10-22, and possibly codons 44-51) might possibly have been introduced into the pseudogene as a unit as a consequence of a gene conversion. I have no suggestion about where these gene segments that appear to have been introduced may have come from initially. It is important to note also that none of the twenty pseudogenes have any unusual similarity with any of the other pseudogenes of the same species or of other species.
I believe, therefore, that the data do not necessarily support the conclusion of Evans and Scarpulla that the nine human pseudogenes were all derived from a consensus of nonprimate mammalian cyto cs genes. However, the statement by Evans and Scarpulla that the nine HC pseudogenes likely originated from an ancestral form of the HCS gene still appears to be valid. How ancient that ancestral form may be, cannot easily be established when sequence changes in pseudogenes involve insertions, deletions and possibly gene conversion events. At the same time, I must add that no one at present knows the role, if any, of these pseudogenes in the mammalian genome. When one examines the data in Table 2 closely, it is evident that dramatic changes would be required for most of these pseudogenes to ever be used for production of functional cytochrome c molecules. On the other hand, is there a possibility that segments of these pseudogenes could be used as a reservoir of genetic information for the production or modification of other proteins? At present that possibility seems unlikely, but it is clearly not impossible. The comment of Neel23 in his Presidential address to the Sixth International Congress of Human Genetics is pertinent in this regard:
In viewing the DNA which is in excess of the needs we can visualize for it as "junk," a hodgepodge of genes rendered useless by evolutionary advance, we have been engaging in an exercise of considerable arrogance.
We should keep our humility and wonder, and with Neel, consider ourselves "privileged to witness and contribute to an unfolding story whose final implications we can only dimly foresee."24
One of the cornerstones of molecular evolutionary theory has been that proteins (and hence genes) have changed progressively through time as one ascends the phylogenetic tree. This author in 196825 raised the following question in regard to protein sequences: if these sequence changes have occurred in the past, they presumably are occurring now. Why is there not marked heterogeneity in protein structure in each species that would show protein sequences in intermediate stages of change? Granted that there is some heterogeneity, but it is clearly not extensive. Also, it has generally been accepted that all intermediate stages of proteins or genes would have to be at least minimally functional. Does the finding of pseudogenes change the answer to the questions posed above? If the pseudogenes noted in humans, rats and mice retained functional coding sequences and controls, then we might consider them as possible candidates for functional genes in the distant future. Since most of the pseudogenes are altered so markedly, the possibility of future function (at least as the same gene) still seems remote. This new knowledge of the structure of pseudogenes then provides additional support for the view that whatever major changes may have occurred throughout evolutionary history, these changes have been a consequence of design, even though the initiation of these design changes may have been triggered by chance events.
Evaluation of Similarity and Homology in Cytochrome c Genes
Now that we have examined some of the experimental data regarding cytochrome c genes and pseudogenes, let us return to some of the questions posed in the introduction, particularly in regard to similarity and homology. First, let us compare the somatic proteins (cyto cs) and genes for these proteins in rats and mice. Based on evidence from paleontology, rats and mice are believed to have diverged from a common rodent ancestor about 30 million years ago. The amino acid sequence similarity for the two cyto cs proteins of rats and mice is 100%, the nucleotide sequence similarity for the coding region of the two cyto cs genes is 97.5%; and nucleotide sequence similarities in the 5'-noncoding region, the 3'-noncoding region and the coding region intron are 86%, 91% and 84%, respectively. I believe, therefore, that the evidence is quite strong that rat cyto cs and mouse cyto cs genes and proteins are homologous, thus supporting the view that there was a common rodent ancestral gene and protein. If we make similar comparisons for rat and mouse cyto ct proteins and genes, the similarities are not quite so striking. The amino acid similarities are 96%; the nucleotide coding region similarity is 93%; while for the 5'-noncoding region and the 3'-noncoding region the corresponding values are 92% and 65%, respectively. However, it should be noted that only a short portion (105 nucleotides) of the 3'-noncoding region was available for comparison. Whether this low similarity value is indicative of the entire 3'-noncoding region is not known. Also no data was available to compare coding region introns of the two rodent cyto ct genes. Although one may presume that the two cyto ct genes are both derived from an ancestral rodent gene, additional data are needed to clearly establish that relationship. It is also clear from the above comparisons that from the time of divergence of rats and mice, the cyto ct genes have diverged more rapidly than cyto cs genes.
However, when we make the corresponding comparisons of the rodent cyto ct proteins and gene with rodent cyto cs proteins and genes, the following values for similarity are obtained: amino acid sequence similarity, 86%; nucleotide sequence similarity of the coding region, 76%; nucleotide sequence similarity of the 5'-noncoding region, the 3'-noncoding region, and the coding region intron, no similarity. In this comparison, although the postulation of a gene duplication as the mode of origin of the cyto ct gene may appear plausible, the present evidence is insufficient to support that hypothesis. Consequently, we cannot say at the present time that genes for the two isozymes of cytochrome c are homologous. At present, the question regarding the origin of the cyto ct gene must remain unanswered.26 It is evident that more studies are needed to see whether the cyto ct gene is present in a wide variety of species, including additional classes other than mammals. Also, if the gene is found in testes of other species, is it expressed as a functional protein in sperm? More complete sequences in noncoding regions and in introns might provide clues regarding the extent of change in control regions among various species, genera, orders and classes of organisms. This again might provide insight into the question of whether the possibility of change in these control or regulatory portions of genes is built into the initial design of an organism's genome (although possibly triggered by chance events), or whether changes are entirely a consequence of sequential chance events (mutations, gene conversions, etc.).
There are some interesting questions regarding the origin of the minor isozymes of cytochrome c found in fruit fly and in yeast (see the nucleotide matrix, Figure 3). In each case, the gene for the minor enzyme (DC3 or CYC7) is very divergent from the gene for the major enzyme (DC4 and CYC1). Did the minor enzyme genes arise by gene duplication? If so, what path did the divergence follow to arrive at the present sequence? I will simply note the problem here, since space will not permit me to deal with questions regarding these two minor cytochrome c isozymes.
Theological Relationships
As this author has previously noted,27 differences in presuppositions may markedly alter questions one may ask regarding origin events. If one accepts the presupposition "that everything may be explained by natural processes," then one must explain how each gene was formed from an ancestral gene. The ancestral gene in turn would have to be relatively simple, since it would have to be formed by chance events from simple precursors. There has been no reasonable scientific proposal for the formation of significant new genetic information (e.g., a gene for a 100 amino acid protein molecule) without the postulation of an intelligent cause. As noted previously,28 postulating that a protein (e.g., cytochrome c) would form by chance is not scientific, when the probability of chance formation is 2 x 10-65. In contrast, if one is open to the presupposition of an intelligent cause, one is free to consider the possibility that ancestral genes may initially have been reasonably complex, even containing appropriate regulatory sequences.
As one examines organisms on any phylogenetic tree, it is clearly evident that many simple organisms (e.g., bacteria) contain genetic information that is not present in mammals. More importantly, mammals contain a great deal of genetic information that is not present in simple one-celled organisms. It is not too difficult to postulate how genetic information may be lost, but it is far more difficult to explain how new genetic information may arise. Gene duplication is most often postulated, but even with gene duplication, the information, although duplicated, is not new. Transfer of gene segments (gene crossovers) may have a role in the formation of some gene families, but again this does not provide new genetic information. Viral genetic information may clearly be incorporated into genes at all phyletic levels, but again this is not new genetic information. I would not attempt to explain how new genetic information may have arisen at various levels of organisms, but would simply postulate that genetic information is present as a consequence of an intelligent cause.
Is it possible that at some level (phyla, classes, orders, genera?), the genetic information is present but not expressed in an ancestral organism, and that this genetic information might subsequently be expressed and processes be initiated leading to species diversity? Could the trigger to initiate the expression of these repressed genes possibly be a chance mutation of a regulatory sequence in a gene that might occur once in a million or more years? The genetic information then expressed might then account for significant changes in organisms. This type of postulation, although reasonable, may prove to be incorrect, but it does illustrate the different type of reasoning possible if one considers not only chance causal events, but also the possibility of an intelligent cause.
In the present paper, the author has used genes and pseudogenes of cytochrome c as models for consideration of some of the most fundamental questions of biology. Although the relationship between these studies and origin questions may not always be clear, it is hoped that this paper will provide the reader with some insight into current areas of investigation that have theological implications. In the author's opinion, scientific research involving the question of origins, including the origin of informational molecules, has been unnecessarily restricted because of the nearly universal acceptance of the view that only chance events must be considered as possible scientific explanations of origin questions.
Whatever the explanation for the origin of genetic information may be, and whatever the roles of design and chance, it is the author's conviction that God is sovereign over all.
©1992
NOTES
1 Reeck, G.R., de Haen, C., Teller, D.C., Doolittle, R.F., Fitch, W.M., Dickerson, R.E., Chambon, P., McLachlan, A.D., Margoliash, E., Jukes, T.H., and Zuckerkandl, E. (1987). "'Homology' in Proteins and Nucleic Acids: A Terminology Muddle and a Way Out of It." Cell 50:667.
2 Mills, G.C. (1990). "Presuppositions of Science as Related to Origins." Perspectives Sci. Christian Faith 42:155-161.
3 Hempsey, D.M., Das, G. and Sherman, F. (1986). "Amino Acid Replacements in Iso-1-cytochrome c." J. Biol. Chem. 261:3259-3271.
4 Mills, G.C. (1975). "Hemoglobin Structure and the Biogenesis of Proteins. Part 1. Relation of Structure to Function for Mutant Hemoglobins." J. Amer. Sci. Affil. 27:33-38.
5 Mills, G.C. (1975). "Hemoglobin Structure and the Biogenesis of Proteins. Part 2. Significance of Protein Structure to the Biogenesis of Life." J. Amer. Sci. Affil. 27:79-82.
6 Mills, G.C. "The role of the Components of the Translation System in Information Transfer." In the proceedings of the conference Information Content of DNA. In press.
7 Scarpulla, R.C., Agne, K.M. and Wu, R. (1981). "Isolation and Structure of a Rat Cytochrome c Gene." J. Biol. Chem. 256:6480-6486.
8 Limbach, K.J. and Wu, R. (1983). "Isolation and Characterization of Two Alleles of the Chicken Cytochrome c Gene." Nucl. Acids Res. 11:8931-8950.
9 Limbach, K.J. and Wu, R. (1985). "Characterization of a Mouse Somatic Cytochrome c Gene and Three Cytochrome c Pseudogenes." Nucl. Acids Res. 13:617-630.
10 Limbach, K.J. and Wu, R. (1985). "Characterization of Two Drosophila Melanogaster Cytochrome c Genes and Their Transcripts." Nucl. Acids Res. 13:631-644.
11 Scarpulla, R.C. (1984). "Processed Pseudogenes for Rat Cytochrome c Are Preferentially Derived From One of Three Alternate mRNAs." Mol. Cell. Biol. 4:2279-2288.
12 Virbasius, J.V. and Scarpulla, R.C. (1988). "Structure and Expression of Rodent Genes Encoding the Testes-specific Cytochrome c." J. Biol. Chem. 263:6791-6796.
13 Evans, M.J. and Scarpulla, R.C., (1988). "The Human Somatic Cytochrome c Gene: Two Classes of Processed Pseudogenes Demarcate a Period of Rapid Molecular Evolution." Proc. Natl. Acad. Sci. USA 85:9625-9629.
14 Evans, M.J. and Scarpulla, R.C. (1989). "Interaction of Nuclear Factors With Multiple Sites in the Somatic Cytochrome c Promoter." J. Biol. Chem. 264:14361-14368.
15 Mills, G.C. (1991). "Cytochrome c: Gene Structure, Homology and Ancestral Relationships." J. Theor. Biol. 152:177-190. Additional references related to cytochrome c can be found in this paper.
16 See note 12 above.
17 See note 13 above.
18 See note 15 above.
19 See note 15 above.
20 See note 2 above.
21 See note 13 above.
22 See note 13 above.
23 Neel, J.V. (1982). "The Wonder of Our Presence Here: A Commentary on the Evolution and Maintenance of Human Diversity." Perspectives Biol. Med. 25:518-558; p. 547.
24 Ibid., p. 551.
25 Mills, G.C. (1968). "The Evolutionary Significance of the Species Variation in Cytochrome c Structure." J. Amer. Sci. Affil. 20:52-54.
26 See note 15 above.
27 See note 2 above.
28 See note 2 above.
GLOSSARY
Coding region of a gene: The portion of a gene that provides the sequence information for the formation of a specific protein.
Codon: The three-nucleotide segment of a gene that codes for a particular amino acid.
Control or regulatory sequence: A sequence of nucleotides in a gene, but outside of the coding region, that controls the expression of the gene; usually 5-15 nucleotides long.
Expressed gene: A gene whose coding region information is expressed in the formation of a specific protein. It is contrasted with a repressed gene.
Eukaryotes: Organisms that have nucleated cells, including yeasts and all higher organisms.
Genome: The total of DNA informational molecules in the cell nucleus of an organism.
Heme prosthetic group: The iron protoporphyrin that provides the catalytic group that permits the cytochrome c protein to function as an electron carrier.
Intron: An intervening DNA segment within the gene that is precisely excised following transcription prior to use of the mRNA for protein synthesis.
Isozymes: Two different but similar proteins that perform the same enzymatic function. In most cases, they have appreciable amino acid sequence similarity.
Non-Coding regions of genes: Those portions of a gene at either end of a coding region (5' end or 3' end) that are transcribed but do not provide sequence information for a protein. They do contain control or regulatory sequences.
Nucleotide: A purine (adenine or guanine) or pyrimidine (cytosine, uracil or thymine) linked to a sugar (ribose or deoxyribose) which is linked to a phosphate.
Pseudogenes: DNA segments in the cell nucleus with nucleotide sequences similar to coding sequences of known functional genes. They are not expressed as proteins. Processed pseudogenes do not have introns or most non-coding control sequences.
Respiratory chain: The group of enzymes which act as a catalyst to the transfer of electrons from a donor compound to oxygen with trapping of energy in the form of ATP; found in all cells with aerobic metabolism.
tRNA and mRNA: Transfer RNA (ribonucleic acid) and messenger RNA; both are involved in the translation of genetic information from a sequence of nucleotides to a sequence of amino acids (protein formation).
Somatic genes: Genes that are present and expressed in all nucleated cells of body tissues. These contrast with tissue-specific genes that are expressed only in certain tissues; for example, testes.